The Gradient Curves Method: An Improved Strategy for the Derivation of Molecular Mechanics Valence Force Fields from ab Initio Data
Abstract
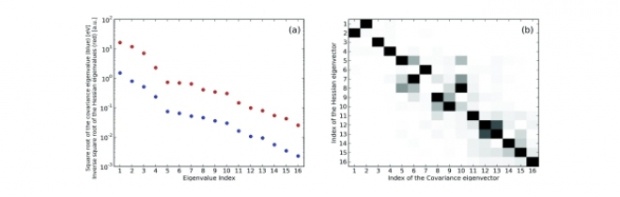
A novel force-field development strategy is proposed that tackles the well-known difficulty of parameter correlations arising in a conventional least-squares optimization. In the first step of the new gradient curves method (GCM), continuity criteria are imposed to transform the raw multidimensional ab initio training data to distinct sets of one-dimensional data, each associated with an individual energy term. In the second step, the transformed data suggest suitable analytical expressions, and the parameters in these expressions are fitted to the transformed data; that is, one does not have to postulate a priori analytical expressions for the force-field energy terms. This approach facilitates the derivation of valence terms. Benchmarks have been performed on a set of small molecules. The results show that the new method yields physically acceptable energy terms exactly when a conventional parametrization would suffer from parameter correlations, that is, when an increasing number of redundant internal coordinates is used in the force-field model. The generic treatment of parameter correlations in the proposed method facilitates an intuitive physical interpretation of the individual terms in the force-field expression, which is a prerequisite for the transferability of force-field models.


![Atomic Velocity Projection Method: A New Analysis Method for Vibrational Spectra [...]](http://molmod.ugent.be/sites/default/files/styles/large/public/ERC_atomic.jpg?itok=qnPTheyw)