Experimental and theoretical IR study of methanol and ethanol conversion over H-SAPO-34
Abstract
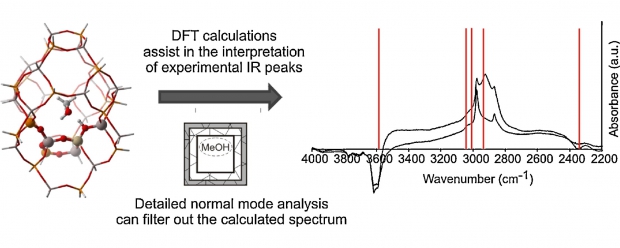
Theoretical and experimental IR data are combined to gain insight into the methanol and ethanol conversion over an acidic H-SAPO-34 catalyst. The theoretical simulations use a large finite cluster and the initial physisorption energy of both alcohols is calculated. Dispersive contributions turn out to be vital and ethanol adsorbs stronger than methanol with approximately 14 kJ mol(-1). Calculated IR spectra of the alcohols and of formed aromatic cations upon conversion are also analyzed and support the peak assignment of the experimental in situ DRIFT spectra, in particular for the C-H and C=C regions. Theoretical IR spectra of the gas phase compounds are compared with those of the molecules loaded in a SAPO cluster and the observed shifts of the peak positions are discussed. To get a better understanding of these framework-guest interactions, a new theoretical procedure is proposed based on a normal mode analysis. A cumulative overlap function is defined and enables the characterization of individual peaks as well as induced frequency shifts upon adsorption. (C) 2010 Elsevier B. V. All rights reserved.
 Open Access version available at UGent repository
Open Access version available at UGent repository