Machines learning MOFs: training time-lagged autoencoders to learn collective variables for accurate free energy surfaces and transition kinetics
Machines learning MOFs: training time-lagged autoencoders to learn collective variables for accurate free energy surfaces and transition kinetics
Promotor(en): V. Van Speybroeck, L. Vanduyfhuys /28217 / Nanoporous materialsBackground and problem
Nanoporous materials have proven promising for applications such as gas storage and separation, nanoshock adsorbers and chemical catalysis. An example are so-called metal-organic frameworks (MOFs) which are hybrid frameworks consisting of inorganic bricks connected to each other through organic. The chemical versatility of MOFs is immense, which also entails a huge engineering challenge as it is practically impossible to synthesize and characterize all possible candidate materials for a certain application. Therefore, we need molecular modeling to accurately describe the behavior of existing materials but also reliably predict the behavior of hypothetical materials. In this respect, one can apply molecular dynamics (MD) to describe the behavior starting from the inter- and intramolecular interactions. However, even with the current computational power available, it still remains unfeasible to describe processes that involve overcoming a large barrier in (free) energy. One example of such processes is given by phase transitions in flexible MOFs where the nanopores evolve from an open large pore phase to a closed narrow pore phase (see figure 1). Another example is given by diffusion of guest molecules adsorbed inside the material from one pore through a narrow window to another pore (see figure 1). In both examples, the initial and final states are characterized by local minima of the free energy or (meta)stable states, while the intermediate structure is given by a saddle point or transition state characterized by a high free energy.
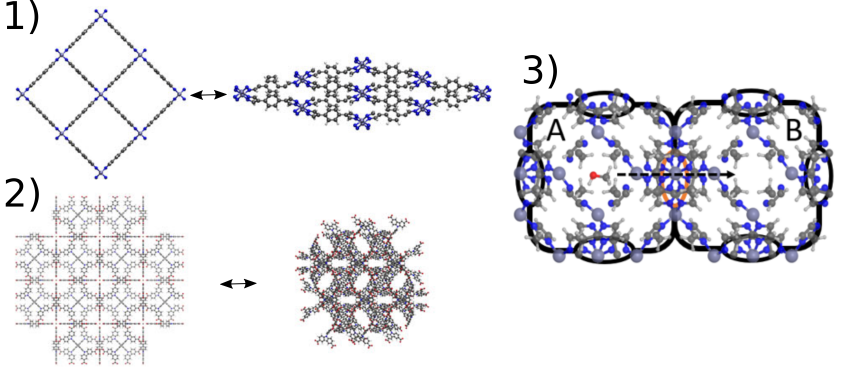
Figure 1: Illustration of phase transitions in flexible MOFs such as 1) CoBDP and 2) DUT-49 as well as diffusion in 3) ZIF-8. Figures taken from [1-2]
Due to the free energy barrier associated with passing through the transition state, these processes occur only very rarely on the time scale accessible in regular MD simulations. Therefore, we need to enhance the simulations, which starts with defining a so-called collective variable (CV), a microscopic degree of freedom that allows to quantitatively describe the progress along the process. During a so-called umbrella simulation, we then enhance the sampling along the direction of this CV by applying a series of biasing potentials in terms of this CV, and reconstruct the free energy profile as function of the CV using statistical physics. However, two important issues remain with this approach: (1) the choice of the CV is mostly done based on intuition which is far from trivial in many cases and (2) to extract the kinetics of the process we need to perform additional restrained simulations in the transition state. In this thesis we aim at addressing both issues more efficiently using machine learning techniques. Therefore, we will consider breathing transitions in CoBDP [1] and DUT-49 [2] (flexible MOFs), as well as diffusion of ethane/ethene through ZIF-8 [3] (MOF with narrow windows) as illustrated in Figure 1. However, depending on the interest of the student, the focus can be set on either one or both issues.
Goal
In this thesis, we aim to tackle both issues outlined above by using a machine learning technique known as time-lagged auto-encoders (TLAE) [4] which is illustrated in Figure 2. Herein, two neural networks are trained simultaneously, one to encode the collective variables in terms of the cartesian input at time t (the encoder), and one to decode the cartesian coordinates at time t+∆t from the collective variables at time t (the decoder).
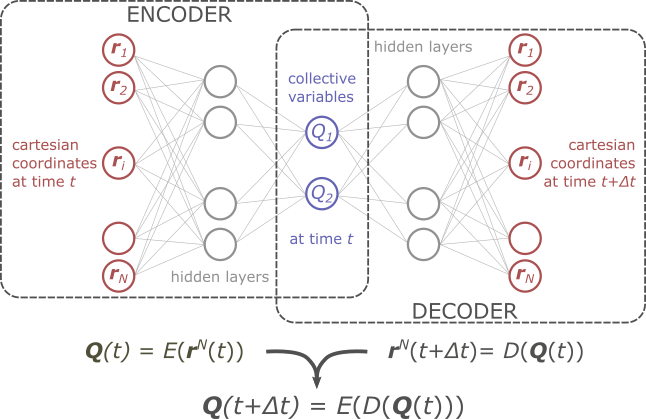
Figure 2: Illustration of a time-lagged autoencoder to simultaneously identify an adequate set of CVs as well as determine their time dependence.
This will allow to determine a set of CVS, Q(r)=[Q1(r),Q2(r)], in which r represents the cartesian coordinates of all atoms in the system, that not only adequately describe the progress along the process, but also adequately discriminate between all relevant intermediate states. Both conditions are required to allow for an accurate construction of the corresponding free energy surface. Furthermore, the TLAE will also be used to determine a relation of the form Q(t+∆t)=K*Q(t) [5] or dQ/dt=f(Q) allowing to simultaneously extract the required information to determine the rate constant from transition state theory (TST) or even going beyond TST.
- Study programmeMaster of Science in Engineering Physics [EMPHYS], Master of Science in Physics and Astronomy [CMFYST]KeywordsStatistical physics, free energy surface, kinetics, Nanoporous materialsReferences
[1] Mason, J. A., Oktawiec, J., et al. (2015). Methane storage in flexible metal–organic frameworks with intrinsic thermal management. In Nature (Vol. 527, Issue 7578, pp. 357–361). Springer Science and Business Media LLC. https://doi.org/10.1038/nature15732
[2] Evans, J. D., Bocquet, L., & Coudert, F.-X. (2016). Origins of Negative Gas Adsorption. In Chem (Vol. 1, Issue 6, pp. 873–886). Elsevier BV. https://doi.org/10.1016/j.chempr.2016.11.004
[3] Verploegh, R. J., Kulkarni, A., et al. (2019). Screening Diffusion of Small Molecules in Flexible Zeolitic Imidazolate Frameworks Using a DFT-Parameterized Force Field. In The Journal of Physical Chemistry C (Vol. 123, Issue 14, pp. 9153–9167). American Chemical Society (ACS). https://doi.org/10.1021/acs.jpcc.9b00733
[4] Wehmeyer, C., & Noé, F. (2018). Time-lagged autoencoders: Deep learning of slow collective variables for molecular kinetics. In The Journal of Chemical Physics (Vol. 148, Issue 24, p. 241703). AIP Publishing. https://doi.org/10.1063/1.5011399
[5] Mardt, A., Pasquali, L., et al, (2018). VAMPnets for deep learning of molecular kinetics. In Nature Communications (Vol. 9, Issue 1). Springer Science and Business Media LLC. https://doi.org/10.1038/s41467-017-02388-1
