Chemical bonds in crystals: a machine learning view
Chemical bonds in crystals: a machine learning view
Promotor(en): S. Cottenier, T. Verstraelen /19MAT05 / Solid-state physicsSince a long time, scientists classify crystals according to the way the atoms in the crystal are connected to each other: rocksalt has ionic bonds, crystalline silicon is covalent, plain iron has metallic bonds, and frozen argon is Van-der-Waals-bound. That’s how you learned it in your introductory courses on the physics and chemistry of solids, and that’s what you’ll find on Wikipedia (https://en.wikipedia.org/wiki/Chemical_bond). However familiar this feels, this classification system has its problems. It originates in macroscopically measurable properties of crystals, such as ‘why does solid argon melt at 84 K and silicon at 1687 K?’, or ‘why is rocksalt brittle and plain iron ductile?’ These criteria were only afterwards connected to the way how atoms bind. They were not designed for the atomic scale in the first place.
In this thesis project, we ask ourselves the question: “how would scientists classify the chemical bonds in crystals if they would start from scratch again, having full atomic-scale information available?” Perhaps, when seen through these eyes, there are other categories possible than ionic or covalent solids. But aren’t our eyes too biased by decades of tradition with the familiar classification system? What if we would ask an unbiased algorithm to look at the data and to make the classification for us?
Goal
This thesis project starts from a data set that has recently been constructed at the Center for Molecular Modeling: for a few hundred binary crystals that span a wide diversity of solids, the three-dimensional charge density difference has been computed by quantum physics methods. This is a 3D picture (or file) that gives for every point in the unit cell the difference between the electron charge density of that point of the crystal, and a superposition of non-interacting free atoms. Therefore, this picture highlights how the free atoms are warped to form the chemical bond. Everything that possibly defines the chemical bond, is encoded in such a picture.
Your task will be to test and apply a series of unsupervised machine learning methods to this picture set. The first step will consist of representation learning: how can the information in the 3D picture be represented in a simpler, lighter way, (almost) without losing information? There are different classes of methods that can be tested: image processing, the use of physical/chemical insight (atom-in-molecule concepts) or deep learning. You will determine the ‘best’ one (most efficient, most accurate, …) The second step is clustering: using machine learning algorithms to group the data representations (crystals) into classes of similar crystals. What is considered to be ‘similar’ depends on the details of the clustering algorithm. If a given method would end up with a clear classification, it is a human task to understand what the chemical reasons are that lead the algorithm to this decision. These classifications can be compared to the results of other quantum physical classification methods, applied to the same set of crystal (electronegativities, electron localization function,…)
Here is your chance to look at solid state chemistry and physics in an entirely new way, and perhaps to set the standards according to which scientists will talk about crystals in the next century. By doing this, you will be introduced in the field of machine learning methods as well.
Fig. 1a+1b: a slice through the difference density for a covalent and a metallic crystal – they are clearly qualitatively different.
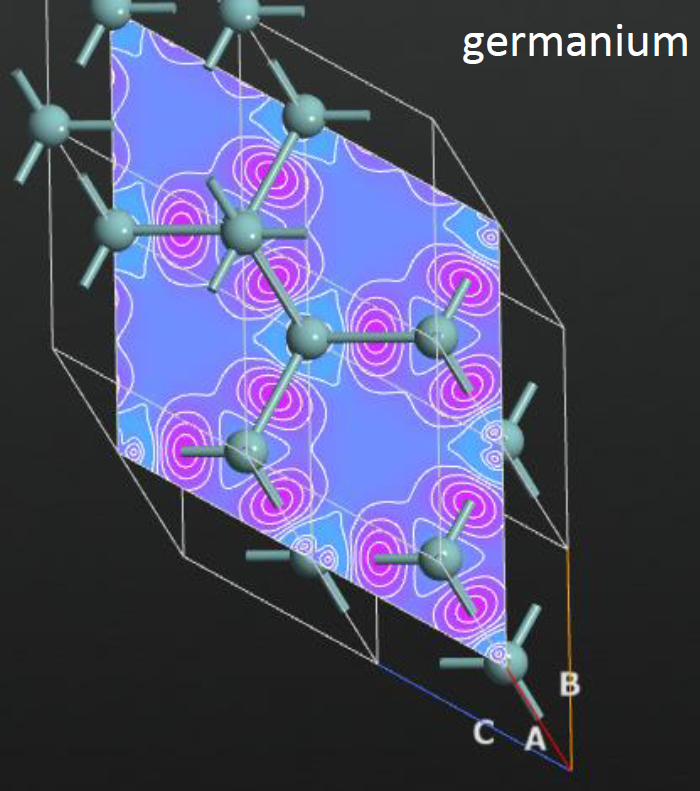

Fig. 1a+1b: a slice through the difference density for a covalent and a metallic crystal – they are clearly qualitatively different.Aspects
Master of Science in Engineering Physics: The nature of interatomic bonding in crystals; Engineering aspects: using machine learning strategies to get insight in complex data sets, applicability to materials optimizationMaster of Science in Sustainable Materials Engineering: Constructing and using materials property databases, structure-property relations via chemical bond interpretation
- Study programmeMaster of Science in Engineering Physics [EMPHYS], Master of Science in Sustainable Materials Engineering [EMMAEN], Master of Science in Physics and Astronomy [CMFYST]ClustersFor Engineering Physics students, this thesis is closely related to the cluster(s) MODELING, MATERIALS, NANOKeywordsmachine learning, Chemical bonding, Crystals


