Drugs as cargo: investigating how peptides can carry drugs through cell membranes using molecular modeling
Drugs as cargo: investigating how peptides can carry drugs through cell membranes using molecular modeling
Promotor(en): A. Ghysels, T.S. van Erp /19MODEV02 / Model and software developmentIn drug design, it is a challenge to bring the less soluble drug molecules into the interior of the cells. A particularly interesting route of thought is to attach the drug molecule as 'cargo' to a cell penetrating peptide (CPP), a short amino acid chain (peptide) that can permeate membranes. Potential clinical applications of this facility have prompted enormous effort, both experimental and theoretical, to better understand how CPPs manage to overcome the prodigious thermodynamic cost of membrane permeation. Current use is limited by a lack of cell specificity, meaning that drugs-loaded CPP cannot distinguish for instance between healthy and cancerous cells. There is currently an insufficient understanding of the modes of their uptake, which hinders developments to improve the specificity e.g. by varying the amino acid sequence, charges, and chain length in a guided manner. Modeling at the molecular scale can aid in understanding the process.
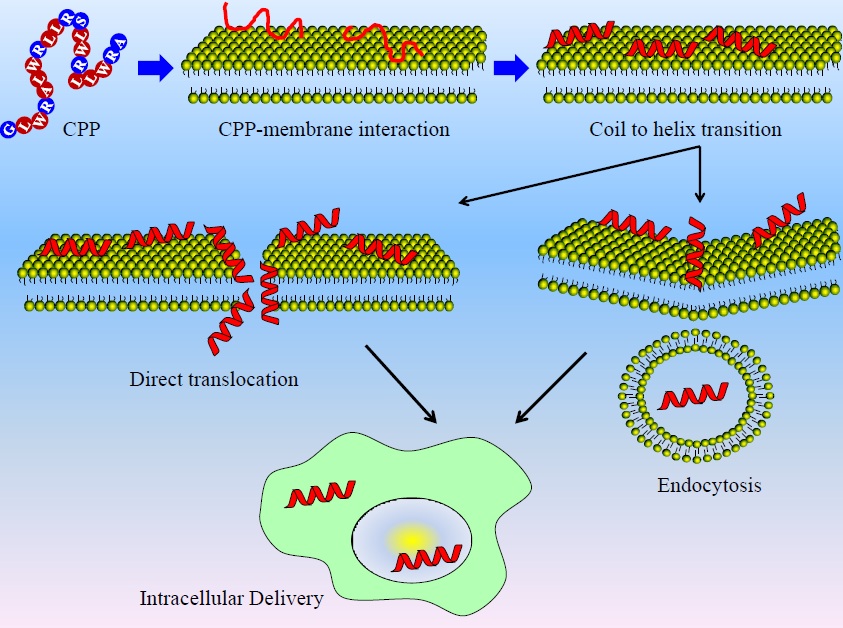
Figure 1. A cell penetrating peptide (CPP) is a short chain of amino acids that can enter the cell and that can carry a drug across the membrane. The route that will be investigated in this thesis is the direct transit through the membrane (translocation).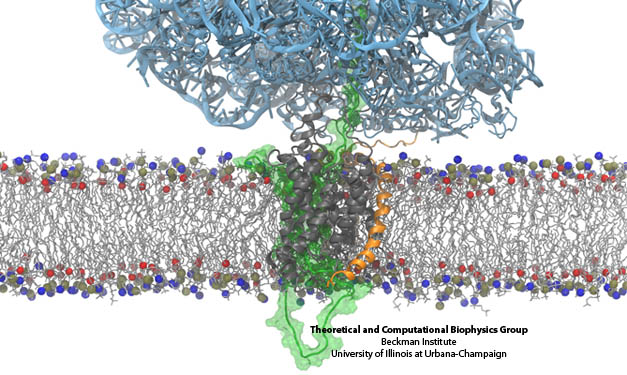
Figure 2. A protein (light blue) is attached to a membrane (horizontal in figure) consisting of phospho-groups (blue and red atoms) and hydrocarbon chains (light grey). Several peptide chains (dark grey and orange) are immersed in the membrane. Water atoms are not shown.Goal
The primary goal of the thesis is to model peptide transport through phospholipid bilayers, also called peptide translocation, in order to understand its process from a biophysics point of view. In particular, the student will tackle the challenge of the time scales. Indeed, when using molecular dynamics simulations to create a trajectory of the peptide, the transition from one side of the membrane to the other might simply not occur during the simulation time, as it is a slow and rare event. Therefore, a new methodology will be developed based on transition path sampling, where multiple trajectories are created by a combination of molecular dynamics and Monte Carlo [1]. The result is a statistical ensemble of many short trajectories of which many are transition trajectories. A re-weighting with probabilities needs to occur to retract the original true rareness of the translocation event.
The above method is called Replica Exchange Transition Interface Sampling (RETIS) and is an advanced method in statistical physics that allows for the extraction of not only thermodynamic equilibrium properties but also kinetics information. RETIS is being tested for small molecules like O2 and H2O at the Center for Molecular Modeling using the PyRetis code that was developed by our Norwegian partner at the Norwegian University of Science and Technology. In addition, the RETIS approach will be held against a recently biasing scheme developed in collaboration with the National Institute of Health. Firstly, a large part of the thesis will be to adapt the current methodology to be suitable for peptide chains that are much larger than O2 or H2O. Secondly, the student will work on understanding the translocation event and its optimal conditions, and on relating the findings to drug delivery applications.
This thesis is embedded in a close collaboration between the Center for Molecular Modeling, the Norwegian University of Science and Technology (NTNU Trondheim), and the National Institutes of Health (NIH, USA).
Aspects
Master of Science in Biomedical Engineering: The modeling of peptide translocation through membranes aids drug design in the view of improved drug delivery.
Master of Science in Engineering Physics: The physics aspect is the fundamental insight in the mechanism of the peptide translocation process and the development of transition path ensembles for extracting kinetics. The engineering aspect is the construction of a methodological simulation tool that aids drug design by attaching drugs to peptides.
- Study programmeMaster of Science in Biomedical Engineering [EMBIEN], Master of Science in Engineering Physics [EMPHYS], Master of Science in Physics and Astronomy [CMFYST]ClustersFor Engineering Physics students, this thesis is closely related to the cluster(s) MODELLING, METHOD DEVELOPMENTKeywordsdrug delivery, method development, Molecular dynamics simulations, membrane permeability, kinetics, biophysicsReferences
[1] Foundations and latest advances in replica exchange transition interface sampling, J. Chem. Phys., 147, 152722 (2017), Raffaela Cabriolu, Kristin M. Skjelbred Refsnes, Peter G. Bolhuis, and Titus S. van Erp
